Maximum covariance analysis (MCA) example
Analyze how temperature across the U.S. covaries
with tropical Pacific sea-surface temperature anomalies
Contents
Load data
clear
load USTA;
latUS = lat;
latmaxUS = latUS(1);
latminUS = latUS(end);
nlatUS = length(latUS);
lonUS = lon;
lonminUS = lonUS(1);
lonmaxUS = lonUS(end);
nlonUS = length(lonUS);
nsUS = nlatUS*nlonUS;
nyear = length(year);
nt = 12*nyear;
TAland = reshape(TA,nsUS,nt);
landUS = land(:);
TAland = TAland(landUS,:);
load SSTPac
landPac = land;
SST = PSSTA(~landPac,:);
SST = SST - mean(SST,2)*ones(1,nt);
latmaxPac = latmax;
latminPac = latmin;
lonmaxPac = lonmax;
lonminPac = lonmin;
nlatPac = (latmaxPac - latminPac)/2 + 1;
nlonPac = (lonmaxPac - lonminPac)/2 + 1;
nsPac = nlatPac*nlonPac;
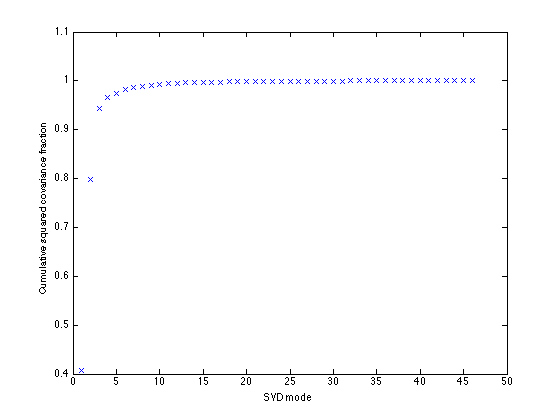
SVD of covariance matrix
Cxy = SST*TAland'/(nt-1);
[U,Sigma,V] = svd(Cxy,0);
s = diag(Sigma);
scf = s.^2/sum(s.^2);
figure(1)
clf
plot(cumsum(scf),'x')
xlabel('SVD mode')
ylabel('Cumulative squared covariance fraction')
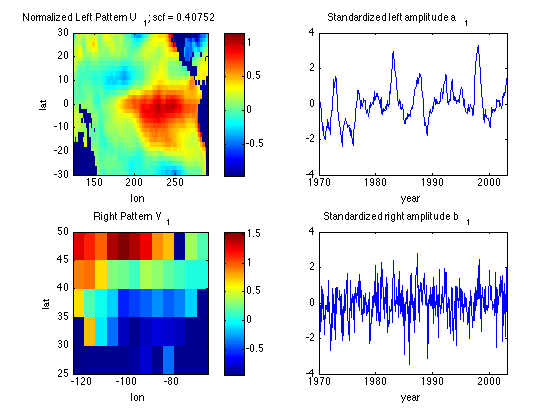
Plot the leading MCA spatial left/right pattern and time series
Normalize by standardizing the time series, so patterns correspond to
a 1 standard-deviation variation in a1 or b1
Also, reverse the sign of U1 and V1 so El Nino SSTA is a positive a1.
Uall = nan(nsPac,1);
Uall(~landPac,:) = -U(:,1);
U1 = reshape(Uall(:,1),nlonPac,nlatPac);
subplot(2,2,2)
a1 = (-U(:,1))'*SST;
plot(yd,a1/std(a1))
xlabel('year')
xlim([1970 2003])
title('Standardized left amplitude a_1')
subplot(2,2,1)
image([lonminPac lonmaxPac],[latminPac latmaxPac],U1'*std(a1),'CDataMapping','scaled')
xlabel('lon')
ylabel('lat')
title(['Normalized Left Pattern U_1; scf = ' num2str(scf(1))])
axis xy
colorbar
Vall = nan(nsUS,1);
Vall(landUS,:) = -V(:,1);
V1 = reshape(Vall(:,1),nlatUS,nlonUS);
subplot(2,2,4)
b1 = (-V(:,1))'*TAland;
plot(yd,b1/std(b1))
xlabel('year')
xlim([1970 2003])
title('Standardized right amplitude b_1')
subplot(2,2,3)
image([lonminUS lonmaxUS],[latmaxUS latminUS],V1*std(b1),'CDataMapping','scaled')
xlabel('lon')
ylabel('lat')
title('Right Pattern V_1')
axis xy
colorbar
sigma1 = s(1)
cov_a1_b1 = a1*b1'/(nt-1)
R1 = corrcoef(a1,b1);
corrcoef_a1_b1 = R1(2,1)
sigma1 =
21.6023
cov_a1_b1 =
21.6023
corrcoef_a1_b1 =
0.2597